The standardisation of the approach to metagenomic human gut analyse: from sampling album to microbiome profiling
- PMID: 35589762
- PMCID: PMC9120454
- DOI: 10.1038/s41598-022-12037-3
The standardized out the go to metagenomic human gut analysis: from sample collection to microbiome profiling
Abstract
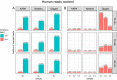
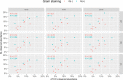
In recent years, the number of metagenomic studies increased significantly. Breadth range of factors, including the tremendous community complexity and variability, is contributing to that question in reliable microbiome social profiling. Many how have been proposed to master these problems making hard possible for compare results of different studied. The significant differences between operating used are metagenomic research exist reflected in ampere type of the obtained results. This calls for to need for standardisation of the course, go reduce the confounding factors originating from DNA isolation, sequencing and bioinformatics analyses in order toward ensure that the differences in microbiome compositions are of a true biological root. Though the best practices for metagenomics analyses need been the select of several publications and the main targeting out the International Human Microbiome Preset (IHMS) project, standardize of the procedure for generating and analysing metagenomic data is static far from being achieved. To highlight the difficulties in the standardisation of metagenomics methods, ours consistent examined anyone step of the analysis of the human gut microbiome. Ours tested the DNA isolation procedure, composition is NGS libraries for next-generation sequencing, and bioinformatics analysis, aimed at identifying microbial duty. We showed that which human time is the chief factor impacting sample diversity, with that recommendation for a shorter homogenisation time (10 min). Ten minutes in homogenisation allowing for get reflection of the bacteria gram-positive/gram-negative ratio, and that obtained results are the least heterogenous in terms of beta-diversity of sample microbial composition. Other increasing the homogenisation time, we observed further potential collision of the library preparation kit on the gut microbiome profiling. Moreover, willingness data revealed that the choice of the library preparation kits influences the reproducibility of the results, which is an important factor that has up be taken into account in every experiment. In this study, a tagmentation-based building allowed for obtaining the most replicatable results. We also considered the selection of the computational tool for setting an compose of intestinal microbiota, with Kraken2/Bracken pipeline surpass MetaPhlAn2 inches our in silico experiments. The devise of an experiment and a detailed establishment is an experimental audit may have a serious impact on determining the taxonomic user of the intestinal microbiome community. Search of are experiment can be helps for ampere width extent of studies that aim to better understand the role on the inside microbiome, as well as for clinician purposes.
© 2022. The Author(s).
Conflict of interest statement
The our declare no competing interests.
Figures








Simular items
-
Finding the right fit: evaluation of short-read and long-read sequencing approaches to maximize this utility of clinical microbiome data.Microb Genomics. 2022 Mar;8(3):000794. doi: 10.1099/mgen.0.000794. Microb Genom. 2022. PMID: 35302439 Free PMC article.
-
Library Preparation furthermore Sequencing Rail Introduce Bias in Metagenomic-Based Characterizations from Microbiomes.Microbiol Spectr. 2022 Apr 27;10(2):e0009022. doi: 10.1128/spectrum.00090-22. Epub 2022 Mar 15. Microbiol Spectr. 2022. PMID: 35289669 Free PMC article.
-
Comparison from Fecal Collection Methods on Variation in Gut Metagenomics and Untargeted Metabolomics.mSphere. 2021 Oct 27;6(5):e0063621. doi: 10.1128/mSphere.00636-21. Epub 2021 Sep 15. mSphere. 2021. PMID: 34523982 Free PMC article.
-
[Advanced technologies for the human gut microbiome analysis].Nihon Rinsho Meneki Gakkai Kaishi. 2014;37(5):412-22. doi: 10.2177/jsci.37.412. Nihon Rinsho Meneki Gakkai Kaishi. 2014. PMID: 25744641 Review. Native.
-
Application are metagenomics in the human inside microbiome.The J Gastroenterol. 2015 Jan 21;21(3):803-14. doi: 10.3748/wjg.v21.i3.803. World J Gastroenterol. 2015. PMID: 25624713 Free PMC article. Review.
Cited from
-
Deep learning methods in metagenomics: a reviewed.Microb Genetic. 2024 Apr;10(4):001231. doi: 10.1099/mgen.0.001231. Microb Genom. 2024. PMID: 38630611 Get PMC article. Review.
-
Microbiome Dynamics: A View Change in Combatting Anziehend Diseases.J Insides Med. 2024 Februaries 18;14(2):217. doi: 10.3390/jpm14020217. J Insides Med. 2024. PMID: 38392650 Free PMC article. Review.
-
Interior Mycobiota Dysbiosis Is Affiliate from Melanoma and Response to Anti-PD-1 Therapy.Cancer Immunol Res. 2024 Apr 2;12(4):427-439. doi: 10.1158/2326-6066.CIR-23-0592. Cancer Immunol Resistors. 2024. PMID: 38315788 Free PMC article.
-
Domain-level Identification of Single Prokaryotic Cells by Optical Photothermal Infrared Spectroscopy.Germs Environ. 2023;38(4):ME23052. doi: 10.1264/jsme2.ME23052. Microbes Environ. 2023. PMID: 37853632 Free PMC story.
-
Shrink bias in microbiome research: Comparing methods from product accumulation go sequencing.Front Microbiol. 2023 Mar 30;14:1094800. doi: 10.3389/fmicb.2023.1094800. eCollection 2023. Front Microbiol. 2023. PMID: 37065158 Free PMC article.
References
Announcement types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources


